CRISPR-Cas9 system is a powerful tool for gene editing, which can target virtually any gene by designing guide RNAs with different sequences. Due to its flexible targeting, high editing efficiency, and ease of operation, it has been widely used in gene function research, genetic engineering, and biomedical fields since its discovery.
Compared to editing single genomic loci, sgRNA libraries typically contain thousands to tens of thousands of sgRNAs, with several sgRNAs per target gene. In CRISPR screening experiments, the library is transfected into target cells and screened based on phenotype, than the genes related to the phenotype are identified by NGS analysis. sgRNA libraries can be designed to target specific categories of genes to the entire genome, without restrictions on sequence, cell type, or species. This allows for high-throughput functional genetic research and rapid and accurate identification of genes related to the phenotype.
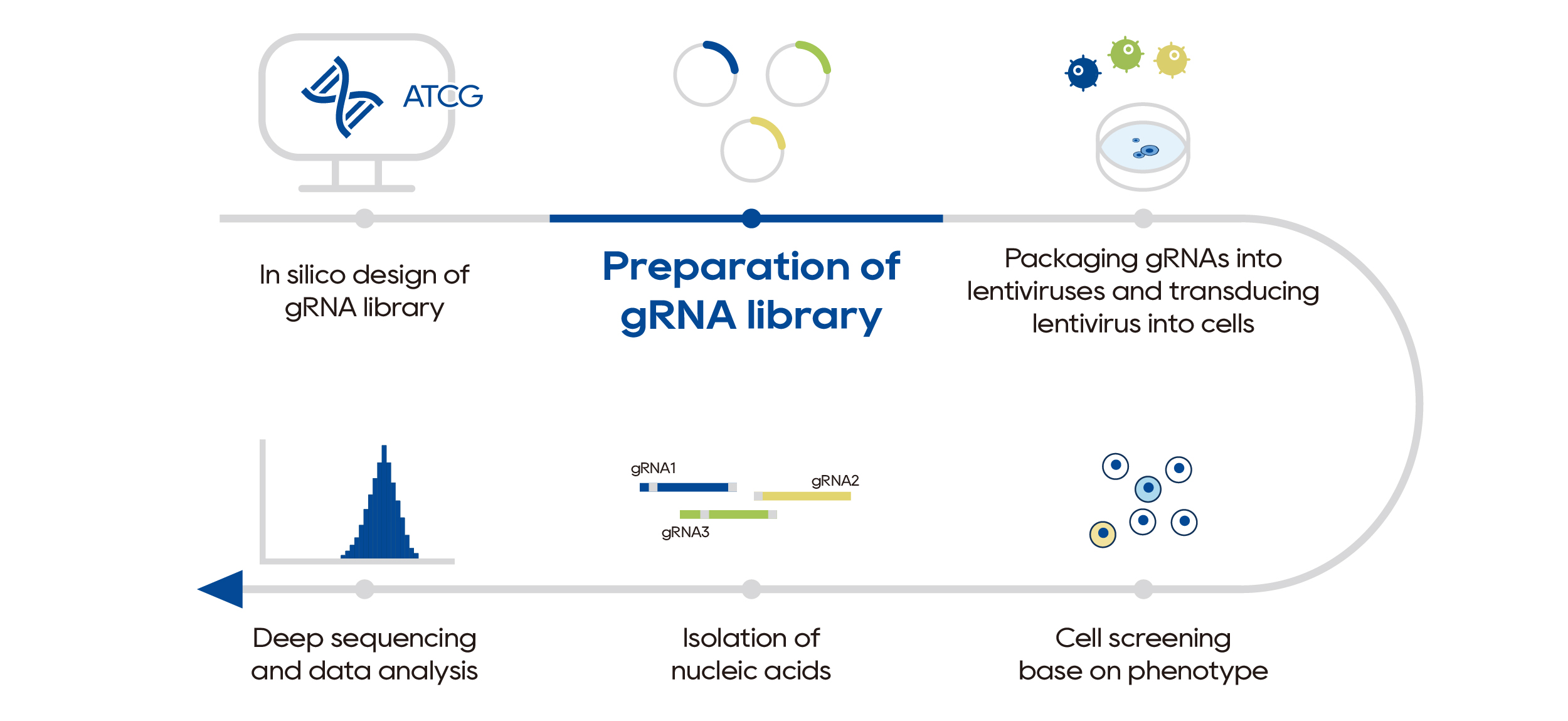
Workflow of functional gene identification
What sgRNA Library can do
1, Functional Genomics Research: By utilizing sgRNA libraries, large-scale gene knockout and functional screening can be conducted to deeply explore gene functions and interactions.
2, Gene Knockout Studies: Through the sequences in sgRNA libraries, knockout studies can be conducted on specific genes or gene families to understand their roles in cell growth, differentiation, metabolism, and other aspects.
3, Phenotype Screening: By utilizing sgRNA libraries, genes related to certain disease phenotypes and traits can be quickly screened out, providing a basis for disease diagnosis and treatment.
4, Drug Development: Through sgRNA libraries, potential drug targets can be screened out, providing candidate targets for new drug development.
5, Genome Editing: sgRNA libraries can be used for genome editing and modification, providing technical support for gene therapy and intervention of genetic diseases.
Custom sgRNA Library
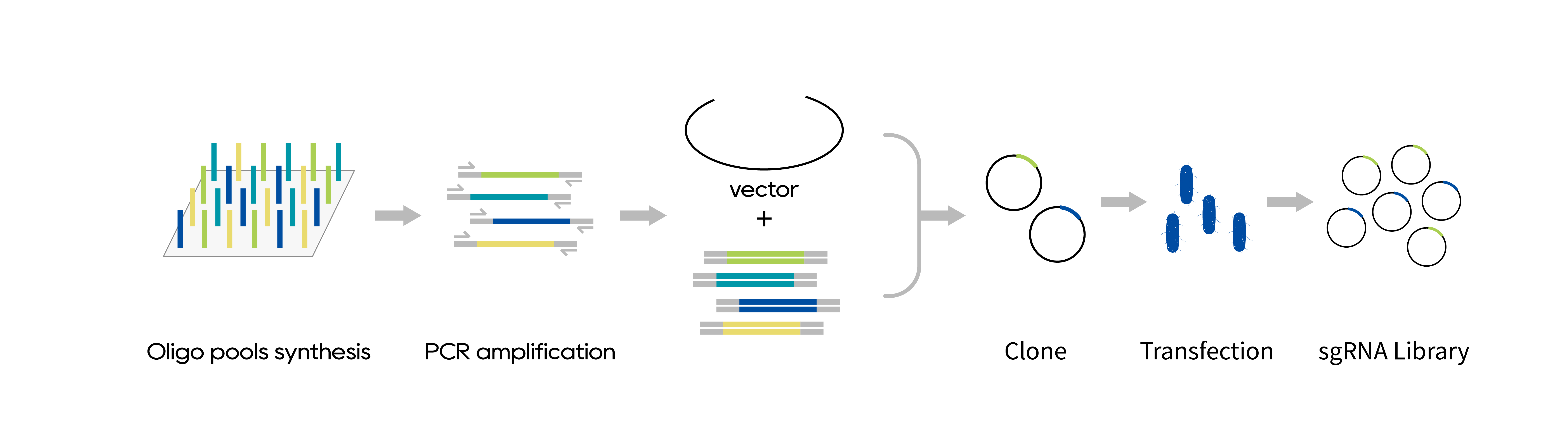
Once the target gene and the corresponding sgRNA sequence are identified, customized oligonucleotide pools are synthesized by the high-throughput synthesis platform. Subsequently, the oligonucleotides are amplified and effectively cloned into the appropriate lentiviral expression vector backbone. The pooled plasmid library can be packaged into lentiviral particles for gene screening in any selected cell line.
Library construction includes oligos synthesis, amplification, and cloning of sequences into plasmids. The delivered plasmids are ready for virus packaging.
 NGSHybridization Capture DNA Probe QuarStar Human All Exon Probes 4.0 (Tumor) QuarStar Human All Exon Probes 4.0 (Standard) QuarStar Liquid Pan-Cancer Panel 3.0 QuarStar Pan-Cancer Lite Panel 3.0 QuarStar Pan-Cancer Fusion Panel 1.0 QuarStar Pan Cancer Panel 1.0 Hybridization Capture RNA Probe QuarXeq Human All Exon Probes 3.0 HRD panel Library Preparation DNA Library Preparation Kit Fragmentation Reagent mRNA Capture Kit rRNA Depletion Kit QuarPro Superfast T4 DNA Ligase Hybridization Capture QuarHyb Super DNA Reagent Kit QuarHyb DNA Plus 2 Reagent Kit QuarHyb DNA Reagent Kit Plus QuarHyb One Reagent Kit QuarHyb Super Reagent Kit Pro Dynegene Adapter Family Dynegene Blocker Family Multiplex PCR QuarMultiple BRCA Amplicon QuarMultiple PCR Capture Kit 2.0 PathoSeq 450 Pathogen Library Corollary Reagent Streptavidin magnetic beads Equipment and Software The iQuars50 NGS Prep System
NGSHybridization Capture DNA Probe QuarStar Human All Exon Probes 4.0 (Tumor) QuarStar Human All Exon Probes 4.0 (Standard) QuarStar Liquid Pan-Cancer Panel 3.0 QuarStar Pan-Cancer Lite Panel 3.0 QuarStar Pan-Cancer Fusion Panel 1.0 QuarStar Pan Cancer Panel 1.0 Hybridization Capture RNA Probe QuarXeq Human All Exon Probes 3.0 HRD panel Library Preparation DNA Library Preparation Kit Fragmentation Reagent mRNA Capture Kit rRNA Depletion Kit QuarPro Superfast T4 DNA Ligase Hybridization Capture QuarHyb Super DNA Reagent Kit QuarHyb DNA Plus 2 Reagent Kit QuarHyb DNA Reagent Kit Plus QuarHyb One Reagent Kit QuarHyb Super Reagent Kit Pro Dynegene Adapter Family Dynegene Blocker Family Multiplex PCR QuarMultiple BRCA Amplicon QuarMultiple PCR Capture Kit 2.0 PathoSeq 450 Pathogen Library Corollary Reagent Streptavidin magnetic beads Equipment and Software The iQuars50 NGS Prep System Primers and Probes
Primers and Probes RNA SynthesissgRNA miRNA siRNA
RNA SynthesissgRNA miRNA siRNA



 Gene Synthesis
Gene Synthesis Oligo Pools
Oligo Pools CRISPR sgRNA Library
CRISPR sgRNA Library Antibody Library
Antibody Library Variant Library
Variant Library







 Tel: 400-017-9077
Tel: 400-017-9077 Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai
Address: Floor 2, Building 5, No. 248 Guanghua Road, Minhang District, Shanghai Email:
Email: 







